CRISPR gene editing technology has many applications in biomedicine and other fields, from the treatment of genetic diseases and cancer, to agricultural breeding, nucleic acid detection and so on. CRISPR gene editing relies on its two components, the guide RNA (gRNA) is responsible for identifying and targeting the target site, and the Cas enzyme is responsible for cutting the target site. CRISPR-Cas9 is the most widely used CRISPR system, but more and more studies have shown that it has potential risks for direct DNA cutting.
In recent years, more and more CRISPR-Cas families have been discovered one after another, among which CRISPR-Cas13 is the shining star among the new CRISPR tools (especially Cas13d). Unlike Cas9, Cas13 is targeted to cleave RNA. The RNA-targeting CRISPR system holds great promise for the development of a new generation of gene-editing therapies. Recently, researchers from New York University and Columbia University published a research paper entitled "Prediction of on-target and off-target activity of CRISPR-Cas13d guide RNAs using deep learning" in the journal Nature Biotechnology.
The research team combined deep learning technology with CRISPR screening to develop an artificial intelligence (AI) platform—TIGER, which can predict the on-target and off-target activities of the CRISPR/Cas13 RNA Editing system, and can also achieve precise regulation of gene expression levels. This new technique paves the way for precise gene regulation in CRISPR gene-editing therapies and further advances the broad applicability of the RNA-targeted CRISPR system in human genetics and drug discovery.
The RNA-targeted CRISPR system has a wide range of application prospects, such as RNA editing, targeted knockdown of mRNA to inhibit specific gene expression, high-throughput screening of drugs, identification of non-coding RNA functions, and can also be used to prevent or treat RNA virus infection. High precision is the key to the safety of therapeutic RNA-targeted CRISPR technology. To advance the clinical application of Cas13, two key goals need to be achieved—maximizing on-target activity and minimizing off-target activity. Off-target activity includes mismatches between the guide RNA and the target RNA, as well as resulting insertions and deletions (indels).
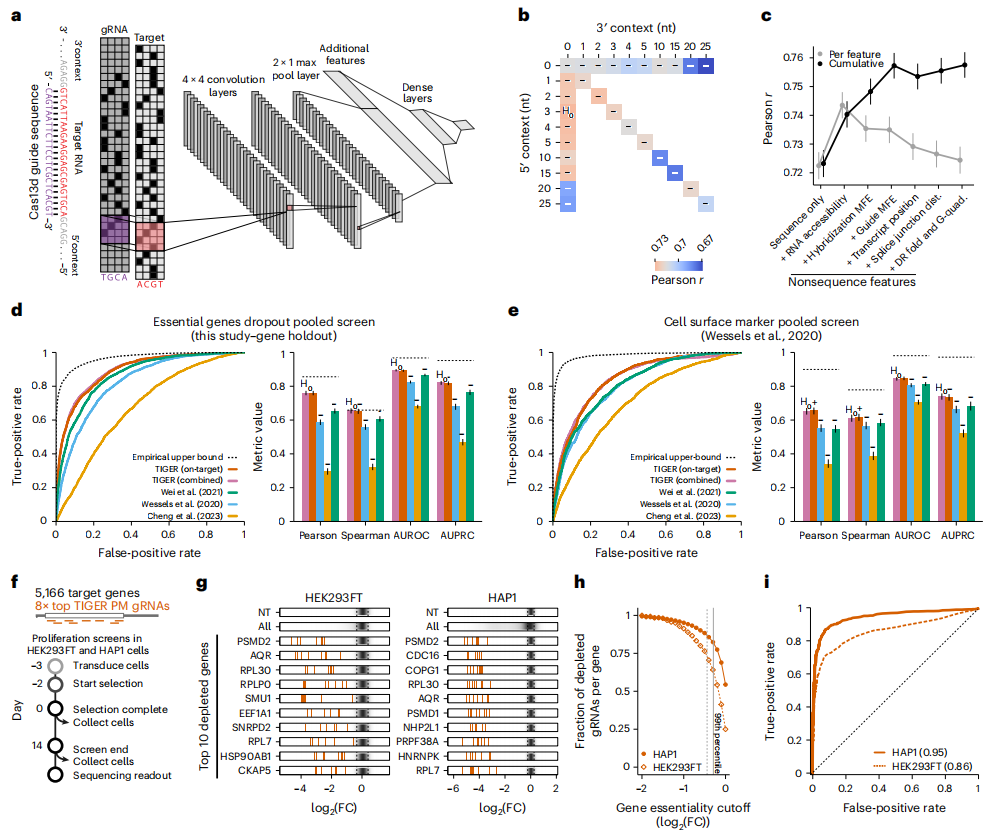
However, early research on RNA-targeted CRISPR systems mainly focused on on-target activity and mismatches, while prediction of off-target activity, especially insertion and deletion mutations, has not been well studied. In humans, approximately one-fifth of genetic mutations are insertion or deletion mutations, so this is an important type of potential off-target to consider in CRISPR design. In this latest paper, Neville Sanjana's team performed a series of RNA-targeted CRISPR screening experiments in human cells, testing the activity of 200,000 gRNAs targeting essential genes in multiple human cell lines, including perfectly matched gRNAs, as well as off-target gRNAs that cause mismatches, insertions, or deletions. A large Cas13d dataset was thus generated for a comprehensive assessment of the on-target and off-target activities of Cas13d gRNAs.
The Neville Sanjana team collaborated with David Knowles, a machine learning expert and an assistant professor of computer science at Columbia University, to train a deep learning model with the above data, and named it TIGER (Targeted Inhibition of Gene Expression via gRNA design). Comparing the results generated by the deep learning model predictions with laboratory tests in human cells, TIGER was able to accurately predict both on-target and off-target activity, making it the first tool to predict off-target activity of an RNA-targeted CIRSPR system.
David Knowles, co-corresponding author of the paper, said that using the huge data sets generated by modern high-throughput experiments, machine learning and deep learning are showing great advantages in the field of genomics. What's more, we were also able to use "interpretable machine learning" to understand why the model predicted the effect of the gRNA so well.
Previous studies in the Neville Sanjana lab have shown how to design Cas13 gRNAs that can knock down specific RNAs, and now with TIGER, it is possible to further guide the design of Cas13 gRNAs, striking a balance between targeted knockdown and avoiding off-target activity. By combining artificial intelligence (AI) with RNA-targeted CRISPR screens, the research team envisions that TIGER's predictions will help avoid unwanted off-target activity, further facilitating the development of next-generation RNA-targeted therapies.
Figure 1. A deep learning model to predict optimal Cas13d gRNAs.
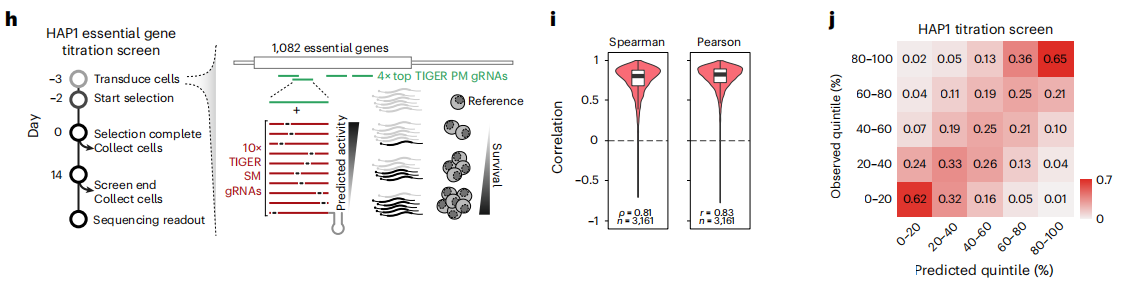
In this latest study, the research team also demonstrated that TIGER's off-target prediction can be used to precisely regulate gene expression levels, and achieve partial expression suppression of specific genes through mismatched gRNAs. This has important implications for many diseases that are caused by increased copy number of genes, such as Down syndrome, certain types of schizophrenia, Charcot-Marie-Tooth disease, and some cancers that are caused by abnormal expression of genes.
Figure 2. Use TIGER to design gRNA to achieve precise regulation of gene expression levels.
Overall, the AI prediction model developed in this study has enhanced our understanding of gRNA targeting specificity and avoiding off-targets, and can also achieve precise regulation of gene expression levels to a certain extent. This research further advances the broad applicability of the RNA-targeted CRISPR system in human genetics and drug discovery.
Reference Wessels H H, et al. Prediction of on-target and off-target activity of CRISPR–Cas13d guide RNAs using deep learning. Nature Biotechnology, 2023: 1-10.