In a new study, researchers from the University of California at Berkeley, Massachusetts General Hospital, Harvard Medical School and the Laurence Berkeley national laboratory identified a key region in Cas9 protein which can determine how CRISPR-Cas9 accurately edited target DNA sequence. Fine-tuning of this domain can produce ultra-precision gene editor and reduce the off-target cutting activity to its lowest level than ever. Relevant research results have been published online on September 20, 2017 in the Journal Nature, with the paper titled "Enhanced proofreading governs CRISPR—Cas9 targeting accuracy". The author is Dr. Jennifer Doudna, professor of the molecular and cell biology in the University of California at Berkeley and the Howard-Hughes Medical Institute researcher.
For CRISPR-Cas9, Cas9 protein has a binding and cutting effect on DNA. Researchers said, the protein region they have identified was a main controller to cut DNA. It would be an obvious target for Cas9 redesign to further improve the cutting precision. This method should help scientists to design custom Cas9 variants, minimizing probability of off-target editing by CRISPR-Cas9. When CRISPR-Cas9 is used for gene therapy in human body, it is a key consideration.
A method to improve the target accuracy could be to introduce mutations into the protein domain called REC3, and then observe which mutations can improve accuracy, while not affect the on-target cutting efficiency.
The another first author, Janice Chen, graduate student from Doudna laboratory said, "we found that even small changes of REC3 domain of Cas9 would affect the difference between the target-editing and the off-target editing, which indicates that this domain is a candidate by inducing mutation to improve targeting specificity. As an extension of this study, we are able to introduce more unbiased mutations into the REC3 domain, compared to our previously targeted ones." The other co-first authors were Yavuz Dagdas in the University of California at Berkeley and Benjamin Kleinstiver of Harvard Medical School.
Ultra-accurate Cas9
In 2012, Doudna and Emmanuelle Charpentier from German Marx-Planck infection biology research institution modified Cas9 protein for other purposes, constructing a cheap and easy-to-use gene editor. Since then, people has sought to reduce the possibility of off-target editing. Although the improved fidelity is conducive to the development of basic research, when the gene editing is going to use for clinical applications, it should be crucial in on-target cutting, because any off-target DNA cutting may make key gene invalid, resulting in permanent and unexpected side effects.
In the past two years, two research teams constructed high-precision Cas9 proteins: eSpCas9, a specifically-enhanced Cas9, and SpCas9-HF1, a high-fidelity Cas9 protein. Chen and Doudna has tried to understand why they are of higher specifically-cutting efficiency than the wild Cas9 proteins from Streptococcus pyogenes.
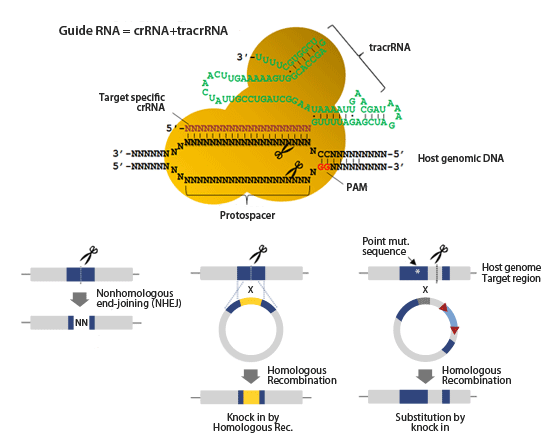
At present, the scientists construct single guide RNA (sgRNA)—a sort of RNA molecules containing 20-nucleotides RNA fragments, and these fragments need to be complementary to the target DNA sequence. The sgRNA would guide Cas9 protein to the target complementary DNA sequence, binding and cutting this DNA region. But this Cas9-sgRNA complex can also bind to other unexpected DNA sites, resulting in unwanted off-target cutting.
In 2015, Doudna laboratory found a conformational switch of Cas9. When sgRNA and target DNA matches, this conformational switch would be activated. They have found that when the sgRNA matches the target DNA precisely, the three-dimensional structure of Cas9, especially its HNH nuclease domain conformation, would change. Thus the cleavage activity of Cas9 can be activated. However, the process that identify the nucleic acids upstream of conformational switch was still unknown.
In the current study, Chen and Dagdas has used a single-molecule FRET (smFRET) technology to accurately measure a variety of protein domains (especially REC3, REC2 and HNH domains) how to move in the Cas9-sgRNA complexes when the complexes bind to target DNAs.
They first identified the specific benefits offered by eSpCas9 (1.1), and SpCas9-HF1, based on these facts that the activation threshold of the HNH conformation switch in these Cas9 variants could be much higher than that in wild Cas9 protein, so that eSpCas9 (1.1) and SpCas9-HF1 variants would not activate the cleavage activity of Cas9 when bind to the off-target sequence.
Then, they found that the REC3 domain could be responsible for detecting accuracy of target binding, and it would instruct the REC2 domain to rotate outward, in order to offer the possibility for HNH nuclease domain change, thereby activating the Cas9 cleavage activity. The Cas9 with such activity conformation would cleave double strand of target DNA.
Chen, Dagdas and Kleinstiver subsequently confirmed that it would be possible to change the specificity of Cas9 protein by making a partial mutation on REC3 fragments, so that the HNH nuclease domain could be not activated, unless the sgRNA and target DNA matches very strictly. They have been able to design an improved ultra-precision Cas9 (named HypaCas9) which retained its target-cutting efficiency, but was slightly better at recognizing target and off-target sites in human cells.
Chen said, "If certain amino acid residues in REC3 mutate, people can adjust the balance of Cas9 between the target cleavage activity and the improved specificity; we can find a balance point to make sure enough cleavage activity at a predetermined target, and reduce off-target events greatly at the same time."
To continuously explore the relationships among structure, function and dynamics of Cas9, Doudna and her team has being devoting themselves to further transform this protein with higher sensitively, so as to use it reliably and efficiently to produce a variety of genetic changes.