Life comes from a single cell (that is oosphere[oosperm]) that repeatedly divides to produce two cells, then four cells, then eight cells, up to about 26 billion cells that make up the newborn. A major challenge in developmental biology is to track how and when these 26 billion cells are produced from a fertilized egg. Until now, there are only snapshots for this developmental process being captured and analyzed in this field.
In a new study, researchers from the Wise Bio-Inspired Engineering Institute in Harward University, Harvard Medical School, University of California, San Diego, and the Sharif Institute of Technology in Iran developed a new method that is possible to make the developmental history tracing come true. Ths[This] approach utilizes a constantly changing genetic barcode to actively record the cell division process of developing mice from each cell lineage in the mouse to its single cell origin, related research results were published in the Journal of Science.
Dr Church, author of the report from Harvard University's Wise Bio-Inspired Engineering Institute and Harvard Medical School, said that, ‘The current pedigree tracking method can only display snapshots of the development process in time, which is due to we have to physically block this development process so that we can observe how the cells look at each stage, just like watching every frame of a movie. But the newly developed method allows us to reconstruct the complete history of each mature cell development which is similar to play a complete movie backwards in real time.’
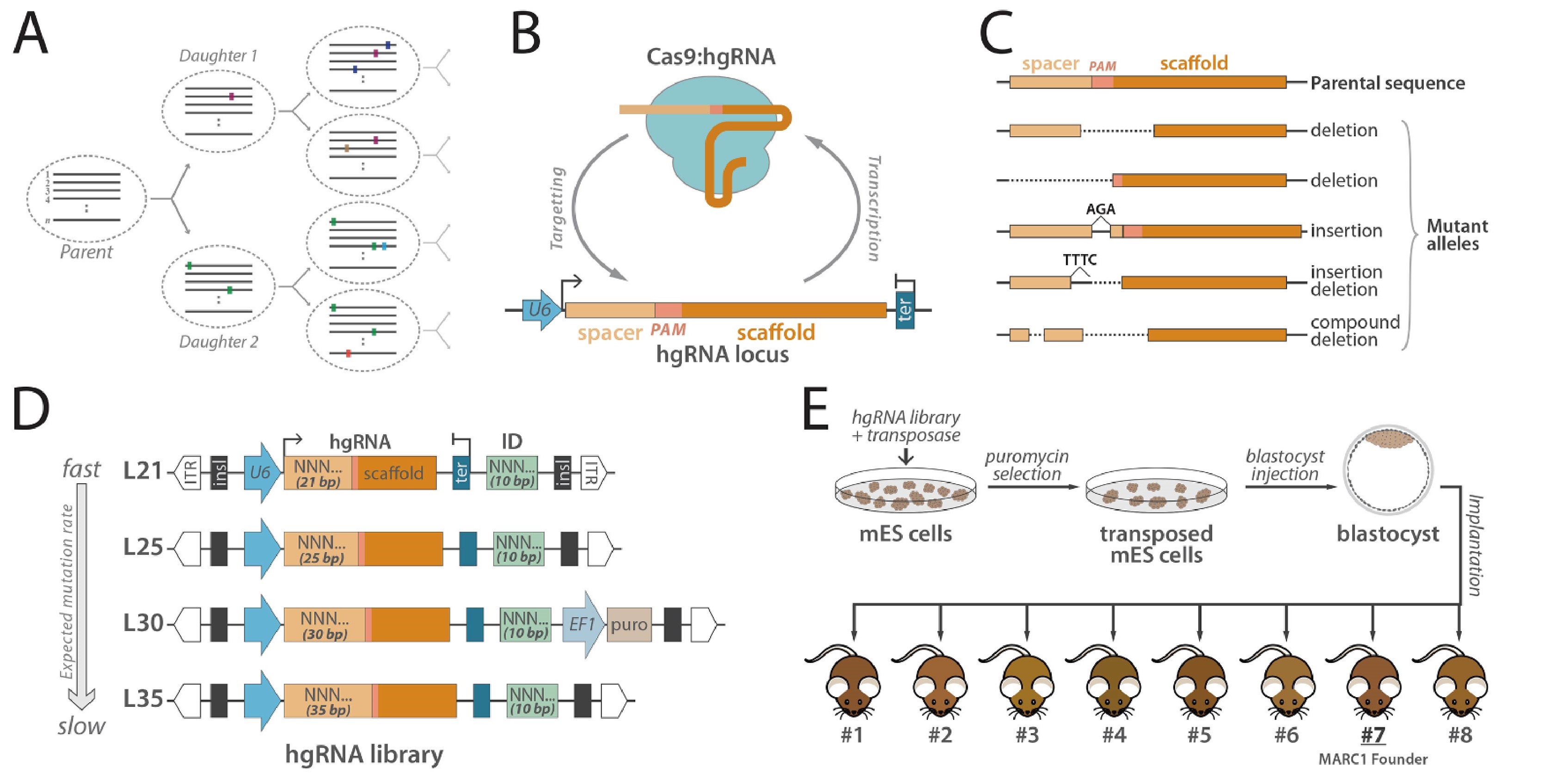
These genetic barcodes were created with a special type of DNA sequence encoding a modified RNA molecule called the homing guide RNA (hgRNA), which was developed by the Church team at the end of 2016 and published in the Journal of Nature Methods. While the enzyme Cas9 is present, the designed hgRNA molecule directs Cas9 to the hgRNA sequence present in the genome, where Cas9 is then cleaved. When a cell repairs this DNA cleavage, it will be able to introduce gene mutations into the hgRNA sequence that, over time, are sufficient to produce a unique barcode.
First, the researchers will develop a ‘founder mouse’ with 60 different hgRNA sequences interspersed in the genome. Afterward, they crossed this initiating mouse with a mouse expressing the Cas9 protein to produce a fertilized egg in which these hgRNA sequences were cleaved and mutated shortly after fertilization.
In vivo barcoding with hgRNAs and strategy to generate mouse with multiple hgRNA integrations (Reza Kalhor et al., Science, 2018).
Dr. Reza Kalhor explained that, ‘In each cell produced by this fertilized egg by division, there will be a possibility of mutation happened to its hgRNA sequence. In each cell division, all daughter cells acquire their own unique mutations in addition to the mutations inherited from the mother cells, so we can track the closeness between different cells by comparing the mutations they have.’
Each hgRNA is capable of producing hundreds of mutant alleles, in general, they are capable of producing unique barcodes that produce a complete lineage for each of approximately 10 billion cells in adult mice.
The continuous record of cell development also allows the researchers to solve a long-standing issue related to embryonic brain tissue, that is, whether the tissue is to distinguish its front end from the back end first or the left side from the right side first. By comparing the hgRNA mutation barcodes present in cells from different parts of the brain of two mice, the researchers found that neurons on the left side of each brain region are more closely associated with neurons on the right side of the same region than neurons from the left side of the adjacent brain region, which indicates that the anterior and posterior divisions of the brain occur earlier than the left and right sides of the brain in the development process of the central nervous system.
Church claimed that, ‘This approach allows us to reconstruct a complete cell lineage tree from the final developmental stage of the model organism until its single-cell stage. It will certainly take many years of hard work to achieve this ambitious goal, but this report represents an important step towards achieving it.’
At present, the researchers are focusing on improving their reading technology so they can analyze the barcode of a single cell and reconstruct the recorded cell lineage tree.
Reference
Reza Kalhor, Kian Kalhor, Leo Mejia et al. Developmental barcoding of the whole mouse via homing CRISPR. Science, Published Online: 09 Aug 2018.